Mycotoxin deactivation with natural enzymes
Using enzymes is one of the efficient approaches against the toxic effects of mycotoxins, which transforms the mycotoxin into a non-toxic molecule. But this approach doesn’t work yet for all types of mycotoxins.
Different mycotoxins have different effects on animals, as they all use different mechanisms. Also some animal species are more sensitive than others when they are exposed to mycotoxins. The mycotoxin deoxynivalenol (DON) for example interferes with the protein production directly at the molecular level. It blocks the peptidyl transferase center of the eukaryotic ribosome. Another mycotoxin, aflatoxin, interferes with DNA replication, but also with DNA transcription. This in turn causes a reduction in the amount of available messenger RNA, which can be rate-limiting for ribosome activity and protein biosynthesis. The effects of other mycotoxins are less direct. For instance, zearalenone, as a mycoestrogen, interferes with endocrine signalling and reduces pig breeding performance. Fumonisins interfere with sphingolipid metabolism and signalling, and low concentrations of fumonisins have an effect on immunomodulation and can increase disease susceptibility. Trichothecenes, in addition to direct interference with protein biosynthesis, also causes reduction of feed intake.
Enzymatic approach
Several approaches and technologies may be used to deal with mycotoxins. These include the plant breeding for more resistance, proper storage conditions or treatment of grains with ozone, ammonia, or alkali. Although none of the treatment technologies have been implemented beyond an experimental or pilot scale. One of approaches, examined and implemented by Biomin, to mycotoxin mitigation is to make additional, microbial enzymes available which act in the gastrointestinal tract and convert mycotoxins to non-toxic metabolites. These enzymes are provided as feed additives. The enzymes can be delivered with their parent microbes. Alternatively, these enzymes are isolated from recombinant production hosts and provided as feed additives in the same manner as other widely used enzymes such as the non-starch polysaccharide (NSP) degrading enzymes or phytases.
Fungal evolution
Fungi have been producing mycotoxins in the environment for many millions of years, and mycotoxins evolved to play a role in natural ecosystems. Biological evolution is driven by interactions. Co-evolution of predator and prey is well-known, and plant pathogen interactions or competition of fungi for substrate, in both of which mycotoxins can play a role, are also examples. Fungal evolution of mycotoxin production elicited evolution of mycotoxin degradation pathways in other microbes. The fumonisin catabolism pathway of Sphingopyxis macrogoltabida MTA144 is a good example. A set of 11 genes, arranged in 1 group in the genome, allows this bacterium to utilise fumonisins as a source of energy and carbon. S. macrogoltabida MTA144 grows faster when a growth medium is supplemented with fumonisins. Expression of fum genes is only turned on when fumonisins are available, and 2 transcriptional regulators encoded in the same cluster of genes are likely responsible for this induction. Homologous genes for catabolism of fumonisins are also present in strain ATCC 55552, and the genes are also clustered, but in a quite different order and orientation. All these facts show that natural evolution has been at work to optimise the selective advantage of bacteria which can break down fumonisins.
2 enzymes of the fumonisin breakdown pathway were studied in detail:
- the fumonisin esterase FumD and
- the aminotransferase FumI.
The substrate selectivity and catalytic activity of these enzymes are a further indication, that the enzymes were optimised for fumonisin catabolism by natural evolution. There are many more examples, where microbial mycotoxin degradation activity has been discovered and described.
Characterisation and production of enzyme
Developing an animal feed detoxification technology based on a microbial mycotoxin degrading enzyme is a comprehensive task with many challenges and possible pitfalls, and therefore, so far only 1 recombinant enzyme has passed the scrutiny of regulatory authorities and reached the market.
One of the key early steps for developing an enzymatic detoxification technology is to establish analytical methods. Quantification of mycotoxin and reaction products must be accurate enough to measure enzyme activity. For filing a dossier with regulatory authorities, the analytical methods also need to be validated. However, in vitro activity in the gastrointestinal tract of animals must also be measured, which required the establishment of biomarkers. Another early significant hurdle of technology development is the ability to produce enough enzyme. A recombinant microbial host strain must be cloned and a matching bioprocess for enzyme production needs to be developed. If the prospects for technology development are promising, the production strain, the fermentation bioprocess, and the enzyme harvesting, drying and formulation process must all be optimised for high-yield and large scale production of active enzyme. The production strain, although it is never released and not contained in the final product, must be accurately characterised for regulatory authorities. A prerequisite for an enzyme to be suitable for detoxification technology is, of course, that the reaction products are known and non-toxic.
Enzymes can be trained
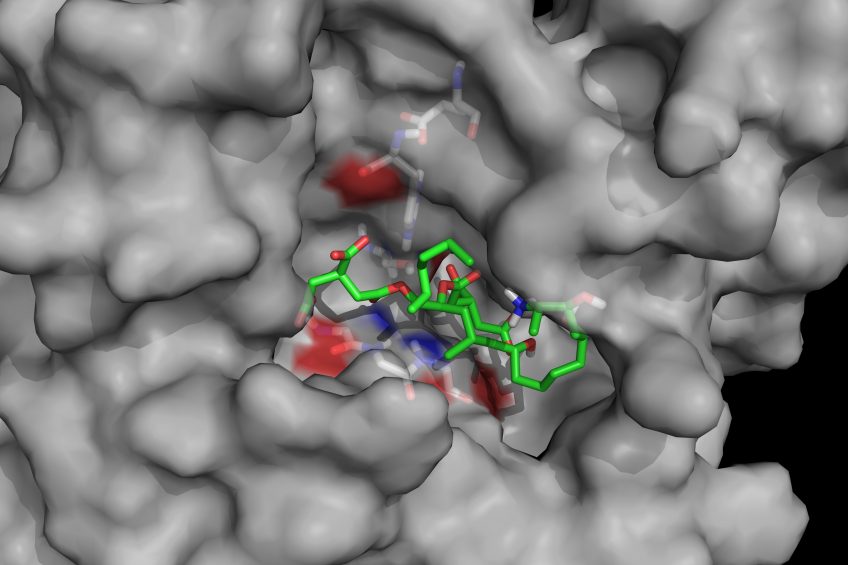
Mycotoxin cleaving enzymes may have gained their selectivity and high catalytic activity through natural evolution but they have not evolved other desirable traits for use as feed additives such as stability during thermal steps of animal feed processing, especially pelleting, or the ability to survive and have high activity in the various compartments of the gastrointestinal tract. Enzymes can, however, be ‘trained’ to have new properties through enzyme engineering. One of the prerequisites of enzyme engineering is an efficient screening system to select superior enzyme variants out of often large pools of variants. Enzyme engineering can be enhanced by knowledge of the 3-dimensional structure of enzymes. When the structure of an enzyme can be predicted by biomolecular modelling, it is possible that the pools of variants that need to be screened can be reduced. Biomin determined the 3-dimensional structures of its enzyme fumonisin esterase FumD and another enzyme by X-ray crystallography.
The future of biotransformation
Adsorption of aflatoxin to binders has been used to make contaminated feed more suitable for animal nutrition. Adsorption to bentonites has been implemented and attempted, with more or less success, for other mycotoxins. Biotransformation is a much more sophisticated development. Reliable and validated biomarkers are important to test the efficacy of the enzymes.
This article was written based on the information of Wulf- Dieter Moll (Biomin Research Center), presented at the 2016 Biomin World Nutrition Forum. References are available on request.













