Sequencing the poultry red mite genome
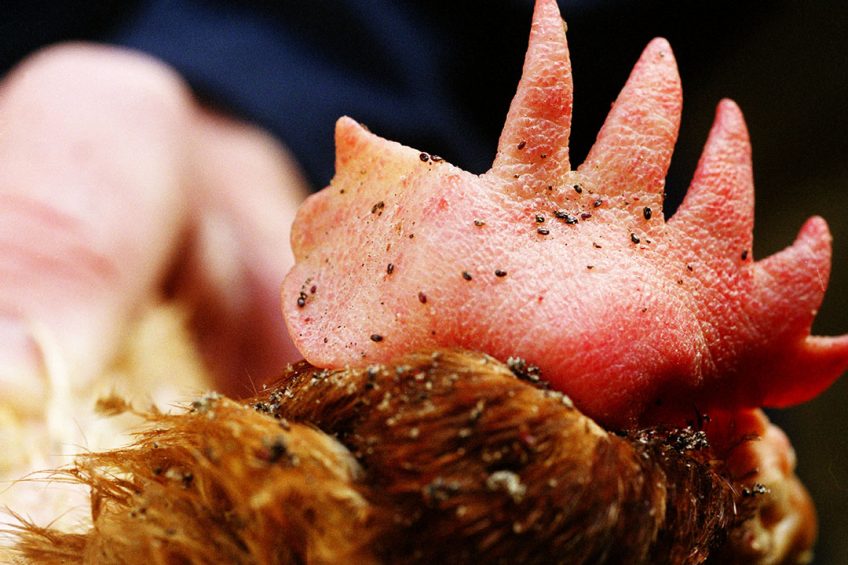
Hopes have risen that the fight against poultry red mite can be won following the successful sequencing of the parasite’s genome.
The red mite, D. gallinae is an ectoparasite of poultry that primarily feeds on the blood of poultry and can spread a large number of diseases between its avian hosts. These diseases cause substantial animal morbidity and mortality as well as huge economic losses for poultry farmers.
Reading the D. gallinae genome
The large array of micro-organisms transmitted by the red mite include Newcastle Disease, avian influenza A virus, Escherichia coli, Pasteurella multocida and Salmonella gallinarum and S. enteritidis.
Now, a team of international scientists led by researchers from the Moredun Research Institute, UK, have provided an insight into how new sequencing technologies enabled them to read the D. gallinae genome. Dr Alasdair Nisbet, who leads the red mite team at Moredun, said researchers had been limited by not knowing the intricacies of the parasite.
“That is why we undertook the genome sequencing project to understand the nitty-gritty of what happens in the mite, what drives the physiological processes and what processes we could interfere with to control it,” he said.
Report author Dr Stewart Burgess said it was the largest mite genome that the team had so far encountered, so the expertise of the international team was vital in bringing the sequencing together into the assembled genome.
“The genome is quite large (960Mb) compared to other closely related mites (typically 60-300Mb). We also do not have access to a clonal or “inbred” population of the mote, which meant that we were potentially working with a polymorphic population.”
Speaking to the American Society for Microbiology, Dr Burgess said that to get round the limitations the team exploited the new third generation sequencing technologies, particularly PacBio and Oxford Nanopore minion sequencing, to make use of the extra-long-read capabilities.
Controlling the parasite
While the size of the genome was larger than initially anticipated, the team is now starting to analyse the horizontal gene transfer and will use the genome to inform aspect of the development of parasitism and the host-parasite interaction in this species.
Dr Burgess said he believed the resource would be a huge asset to those involved in poultry red mite research, as scientists looked to find new ways to control the parasite.
“The genome is still being improved, as we look to incorporate further transcriptomic datasets and also to use additional minion data to further reduce the number of scaffolds in the genome assembly.
“That said, we are currently using the genome to identify SNPs associated with particular proteins that may be useful in mite control and also that might confer acaricide resistance.
“The genome sequencing project now represents a significant tool allowing the design of new methods of control through a greater understanding of the biology of the parasite. It will also be a really valuable tool for molecular epidemiologists trying to understand the source and spread of the parasite in populations of birds.”
Read also: New red mite treatment hits market
Dr Nisbet stressed that at present there was no one thing to control the parasite: “Vaccination will be one of the control methods in the future, but no single thing will be a silver bullet.
“Other strategies will include biosecurity, appropriate treatments at the right time, nutrition and breeding.”
Draft Genome Assembly of the Poultry Red Mite, Dermanyssus gallinae was published late last year in the American Society for Microbiology.












